Element:CSVMV
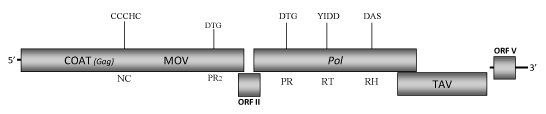
DescriptionCassava vein mosaic virus (CsVMV) is a plant pararetrovirus found primarily in Brazil, where it infecting cassava plants (Manihot spp.) produces mild symptoms characterized by chlorosis followed by mosaic veins, leaf distortion and epinasty of the young leaves (Calvert et al. 1995; de Kochko et al. 1998). CsVMV is the type species of Cavemovirus genus (for more details see ICTVdb) of Caulimoviridae plant virus family and together to the Tobacco vein clearing virus (TVCV) are the two species of Class 3 in the phylogenetic analysis performed by Llorens et al 2009. CsVMV has isometric particles of 50-60 nm in diameter (Kitajima and Costa, 1966) composed of a genomic 8159 bp dsDNA (de Kochko et al. 1998). The genome of CsVMV is organized in five ORFs (de Kochko et al. 1998) and not in six as previously reported (Calvert et al. 1995, U20341). It is due to the presence of two Adenine (AA) (instead of only one A) at position 6817-18 that induce a frameshift such that previous described ORF IV and V become only one ORF (ORF IV) (de Kochko et al. 1998). The ORF I contains both the COAT (gag) and movement protein (MOV) domains in the same order of the other Cavemovirus species TVCV (de Kochko et al. 1998; Calvert et al. 1995; Lockhart et al. 2000). The consensus RNA-binding sequence associated to the putative COAT-gag region, like that of other plant pararetrovirus, is larger than the related retroelement consensus RNA-binding sequence (C-X2-C-X4-H-X4-C) because it includes an additional cysteine (Cys) at the -2 position (C-X-C-X2-C-X4-H-X4-C ) (Hull 1996; Bouhida et al. 1993). ORF III and IV display respectively the pol-polyprotein domain organization (protease, reverse transcriptase and RNaseH) and the transactivator region (TAV) typical of the other caulimoviruses. ORF II (between Gag-MOV and Pol genes) and ORF V (downstream of the intergenic region ) do not encode for any described protein, even if the ORF V-oligopeptide could have a putative role in the transcription regulation (de Kochko et al. 1998). Interestingly, our analyses have revealed the presence of a second protease-like domain downstream to the MOV region (C-terminus of ORF I) that shows sequence similarity to the conventional Pol-protease domain. A similar feature has been also observed in the genome of the two soymoviruses - Peanut chlorotic streak virus (PCSV) and Soybean chlorotic mottle virus (SbCMV ) - and of the tungrovirus Rice tungro bacilliform virus (RTBV), but it is not clear if these sequences are a functionally active proteases additional to those found in the Pol genes. CsVMV can be transmitted by mechanical inoculation, as well as by stem cuttings that is used as reproductive material in the vegetative propagation (Calvert et al. 1995). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||