Description
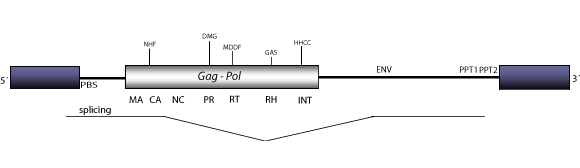
Bagy-2 is a LTR retroelement originally described along with other retrotransposons in the genome of Hordeum vulgare. This element belongs to Athila clade, a lineage of possible retroviruses in plants (Wright and Voytas 2002), which is sibling of Tat clade, a lineage of plant LTR retrotransposons. Both lineages provide evidence of a shared ancestral lineage originally described as Class A by Wright and Voytas (1998). From a study focused on the mechanisms of retroelement-driven genome expansion counteractions, Bagy-2 was established as a continuum of 10 Kb interrupted and arranged as multiple nested insertions within a heterochromatic region. The figure below depicts the consensus structure of Bagy-2 in which both LTRs flank a reconstructed internal region that presents a Primer Binding Site (PBS) complementary to a tRNAAsp, a single Open Reading Frame (ORF) gag-pol, two Polypurine Tracts, PPT1 and PPT2 (Shirasu et al. 2000; Wright and Voytas 2002), and also, an additional env gene which undergoes splicing to generate a subgenomic env product (Vicient, Kalendar and Schulman 2001).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|