Element:BSOLV
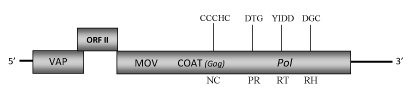
DescriptionBanana streak virus (BSV), the causal agent of viral "leaf streak disease" of banana and plantain (Musa spp.) (Lockhart, 1986), is a member of the plant pararetrovirus genus Badnavirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). Several isolated have been studied, one of which is BSOLV infecting a triploid plantain cultivar “Obino L'Ewai” (used as the female parent in the production of improved tetraploid hybrids, Harper and Hull 1998). According to Llorens et al. 2009, BSOLV belongs to Badnavirus genus within Class 2 of the Caulimoviridae family. The analyzed BSOLV (Nigerian isolate) has bacilliform particles of 30 x 150 nm in size containing a circular double-stranded DNA genome of about 7.3 kb in size (7389 bp long) which three Open reading frames (ORFs I, II, III) potentially coding for proteins (Harper and Hull 1998). The first two ORFs encode two small proteins: the ORF I-product reveals sequence similarity to the virion associated protein (VAP) present in mainly of caulimoviruses, while the function of ORF II is not yet well known. The third large ORF III encodes a polyprotein containing regions with homologies to a movement protein, a gag-RNA binding domain, to an aspartic protease (AP), reverse transcriptase (RT) and RNase H (RH) domains typical of the badnavirus species (Harper and Hull 1998). The plant pararetrovirus consensus RNA-binding sequence associated to the putative gag region is larger than the related retroelement consensus RNA-binding sequence (C-X2-C-X4-H-X4-C) as it includes a large region rich in zinc finger (CCHC) array duplications (only preserved in Badna and Tungroviruses) similarly to those of several LTR retroelement nucleocapsids (Hull 1996; Bouhida et al. 1993; Llorens et al. 2009). The viral "leaf streak disease" was first described in Ivory Coast, Africa in 1968 (Lassoudiere 1974), but now is probably distributed worldwide in cultivated banana and plantain crops. The symptomatology is correlated with variations in virus concentration. The symptoms may vary from faint broken chlorotic lines to necrosis of young leaves, internal necrosis of the pseudostem and plant death. The bunch size and fruit quality are also compromised (Lockhart, 1986). The virus is transmitted naturally by the two mealybug vectors (Pseudococcidae) Planococcus citri and Saccharicoccus sacchari (both of which colonize banana), as well as by seed, even if the vegetative propagation is the principal method of virus spread. Unlike the petuvirus Petunia vein clearing virus (PVCV, Richert-Pöggeler and Shepherd 1997), no obvious integrase motif could be identified in the BSV sequence (Harper and Hull 1998). However, multiple BSV-DNAs were found to be integrated into the nuclear genome of the cultivar (cv) Obino l’Ewaï (Harper et al. 1999; Geering et al. 2005) as well as in that of other Musa cvv. (Ndowora et al. 1999; Harper et al. 1999; Gayral et al. 2008; Gayral and Iskra-Caruana 2009). Two types of BSV-Endogenous pararetrovirus (EPRVs) sequences have been described in banana. The first type sequences, defined as noninfectious (with nonfunctional viral ORFs and/or incomplete viral genomes), are present in the two most common Musa species Musa acuminata (denoted A) and Musa balbisiana (denoted B) that, together to Musa schizocarpa (denoted S) are the wild progenitors of the domesticated banana ( Geering et al. 2005). The second EPRV sequences, so-called infectious type, contain the complete functional viral genome and have been detected in some Musa genotypes (i.e AAB and BB groups, Geering et al. 2005 ; Gayral et al. 2008). In particular, the BSOLV-integrant sequence appeared particularly complex: it consists of two segments (which together constitute the full complement of the virus genome) separated by a "scrambled region" containing non-contiguous and inverted viral sequences (Ndowora et al. 1999; Geering et al. 2005). Interestingly, the absence of episomal BSV in the "mother plants" before the tissue culture procedure and the presence of viral sequences into the propagated material strongly suggest for an integrated origin of the episomal forms detected during the proliferation stage (Dallot et al. 2001; Gayral et al. 2008). Indeed, it has been demonstrated that in vitro micropropagation procedure could play a possible effect on BSV expression in healthy plants (episomal BSV-free) carrying EPRV sequences and that the proliferation stage is clearly determinant in the appearance of episomal BSV (Dallot et al. 2001). The high degree of sequence conservation between the integrated and episomal form of the virus indicates a recent integration event (Gayral et al. 2008). Moreover, recent independent integration events occurred after the divergence of the three banana species (A, B and S), also indicate that viral integration is a recent and frequent phenomenon Gayral and Iskra-Caruana 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||