Element:BIV
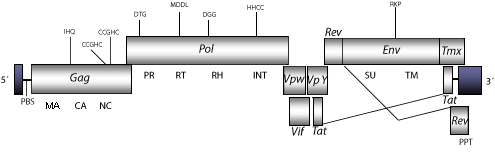
DescriptionBovine Immunodeficiency Virus (BIV) is a lentivirus originally isolated in 1969 in Lousiana from a cow infected with lymphocytosis and lymphoadenopathy (Van Der Maaten et al. 1972). BIV was later associated with disorders such as lower milk production, immunodeficiency, encephalitis, bovine paralysis syndrome, skin disease and weight loss (McNab, Jacobs and Smith 1972; Walder et al. 1995). Two clones (127) and (106) of BIV were sequenced by Garvey et al. (1990). As BIV is found predominantly in monocytes and splenic macrophages in vivo, but not readily infecting T cells, the BIV clone 127 were experimentally used to design a gene transfer system (Gonda et al. 1994; Berkowitz et al. 2001). The full-length BIV consensus is ~8.4 Kb in size including variable LTRs of 696-716 nt (pro5´LTR of 127-201 and pro3´LTR of 495-589 nt). Both LTRs contain regulatory elements and enhancer sequences related to binding sites for the Sp1 and NF-kappa B factors. The genbank accessions M32690 and L04972 have been selected to describe the 5´LTR and 3´LTR, respectively. The internal region displays a Primer Binding Site (PBS) complementary to a tRNALys, Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, tat and rev (both expanded in two exons) and vif, which are three accessory genes common in other lentiviruses, vpw, vpy, and tmx accessory genes, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Garvey et al. 1990; Berkowitz et al. 2001). As other non-primate lentiviruses, BIV displays between the RNase H and INT domains, an ORF coding for a dUTPase, similar to that observed in betaretroviruses, N-terminal to the PR domain (Elder et al. 1992; Payne and Elder 2001 and references therein). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||