Element:BFV
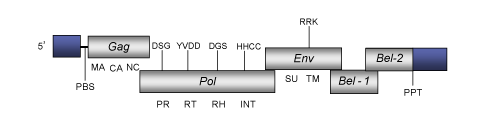
DescriptionBovine foamy virus (BFV) is a foamy retrovirus originally isolated from bovine species. BFV belongs to genus Spumaretrovirus within Retroviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. (2009) is located within Class 3 of the Retroviridae family. The foamy viruses family causes a pronounced cytopathic effect in a wide variety of tissue culture cells, but BFV appears to be benign in vivo. This lack of apparent pathogenicity has led to the idea that the foamy viruses may be suitable delivery vectors for gene therapy (Russell and Miller 1996; Schmidt and Rethwilm 1995). However, it has been suggested that BFV presence in the host may contribute to diseases caused by other pathogens (Marino et al. 1995). The genomic structure of BFV is 12.2 Kb in size including LTRs of 1.305 Kb (Holzschu et al. 1998). The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNALys, three Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, the 3´accessory Bel-1 (BF-ORF1) and Bel-2 (BF-ORF2) genes (Renshaw and Casey 1993), and a Polypurine Tract (PPT) adjacent to the 3´LTR (Holzschu et al. 1998). BFV gag and pol ORFs overlap, and contain the features characteristic of retroviruses (gag, protease, reverse transcriptase, RNase H and integrase) (Holzschu et al. 1998). The env gene encodes a protein that is over 40% identical in sequence to that of Human foamy virus (HFV). Two accessory genes, Bel-1 and Bel-2, are located downstream to env. The first one includes a short overlap between env gene while Bel-2 overlaps with the first accessory ORF and the 3´LTR region terminal (Holzschu et al. 1998). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||