Translational transactivator Protein
The "Transactivator/viroplasmin protein" (TAV) -also known as "Inclusion body matrix protein" (IBMP)- is a multifunctional protein encoded by several caulimoviruses such as those belonging to the Caulimovirus, Soymovirus and Cavemovirus genera and involved in many aspects of the virus life cycle (Haas et al. 2005; Kobayashi and Hohn, 2003; Petrzik et al. 1998; Richins et al. 1987; Hull et al. 1986; Hasegawa et al. 1989; Mushegian et al, 1995; Glasheen et al. 2002;de Kochko et al. 1998; Lockhart et al. 2000).
The Cauliflower mosaic virus (CaMV)-ORF 6 product (P6) is probably the most extensively studied TAV (see also Covey and Hull 1981). In this viral species, TAV has been shown to be essential for basic virus replication (Kobayashi and Hohn, 2003) and it is the major component of CaMV inclusion bodies or viroplasm, which are large membrane-free structures (with the size of organelles) where CaMV protein synthesis, reverse transcription and virion assembly occur (Kobayashi and Hohn, 2004; Haas et al. 2005). TAV has also been shown to promote the translation of the polycistronic CaMV second major transcript 35S RNA (Hohn et al. 1990) and it is capable to induce hypersensitive response (HR) in some plant hosts (Kobayashi and Hohn, 2004). Prior studies demonstrated that CaMV-TAV is the major determinant of host specificity and influences symptom severity (Daubert et al. 1984; Daubert and Routh, 1990; Agama et al. 2002), and it has also been suggested to have post-translational roles in virus replication (Kobayashi and Hohn, 2003).
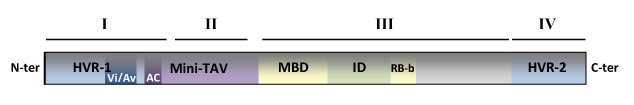
TAV has been considered a mosaic of different domains whose function independently and have different properties (De Tapia et al. 1993; Kobayashi and Hohn 2003; 2004). The figure below shows a schematic representation of TAV domain architecture. Based on Kobayashi and Hohn (2003; 2004) four domains (I, II, III, IV) can be distinguished in TAV. Domains I and IV include two hypervariable regions (HVRs 1-2) that appear to be responsible for host selection and/or pathogenesis. Domain I contains the region required for CaMV virulence and avirulence (Vi/Av) and plays also role in viroplasm formation. Domains II and III contain the "caulimoblock" a preserved region between amino acids (aa) 283 and 350 (De Tapia et al. 1993) that probably play a role in basic virus replication. Domain II includes the "Mini-TAV", a small portion of protein containing the active center (AC) of TAV which interacts with ribosomal protein L18, double-stranded RNA (dsRNA) and RNA–DNA hybrids (Kobayashi and Hohn, 2003; Cerritelli et al. 1998). Domain III is a multifunctional region that includes a multiple binding domain (MBD or RBa; aa 242 to 310) with affinity for both protein and RNA; a second RNA domain (RB-b; aa 346 to 378) as well as an interactive domain (ID) proposed from the competition experiments for transactivation activity (De Tapia et al. 1993; Kobayashi and Hohn, 2003).