Menu Tools: Management
Menu Tools: Management
Management overview
GPRO offers you three distinct tools for performing data mining and managing files, folders and contents within folders. If you make click on the menu tab "Management" a drop-down list will show you appears three tool options; “File and folders”; “Find sequences”; and “Alignments”.
Files and folders
This tool is organized in three sub-sections called "Join Folders", "Join Files" and "Split Files" and that you can access just by clicking on their respective tabs at the top of the GUI.
Join folders
“Join folder” is a script that allows you to reorganize folders using distinct key terms such as enzyme or annotation names, etc (even if folders are in other folders within the Directory). Figure 10.1 shows an example consisting on the following steps:
- 1) Use the mouse to drag the folder containing all the relevant folders from the Directory to the "input folder" text box.
- 2) Type as many “filter words” as needed in the box “By name” below the tag “Filter options” in order to define the criteria for organizing the folders (all folders called with an identical or similar term will be selected). Then, press add and words will pass on to the text square below.
- 3) Check any of the three “Exact term”, “Regular expression” or “Case sensitive” options
- 4) Use the mouse to select an output folder from the directory and drop it to the output box at the bottom of the GUI and make click on the button “Proceed” to run the script, which will select and export all folders fitting to your search term/s in the output.
|
Figure 10.1. Join folders using distinct folders with contents concerning LTR retroelement protein domains as an example : 1) drag folder in analysis to the "input folder" area; 2) type “filter words” and options; 3) use the mouse to select an output folder and drop it to the output box; 4) run the script; 5) the script will export all folders joined in other folders named according to the terms used as as key words. |
Join files
The script “Join files” allows you to select and to group files either in a single file and/or folder. The tool accepts only FASTA and XML files. Using “Join files”, you can collect, for example, the distinct XML files of a BLAST search in a single XML file or to retrieve common gene features of an annotation database divided into distinct files. The possibilities offered by this tool are the following:
- Select files and place them in a single folder
- Select XML files and place them in a single folder
- Join files in a single file
- Join XML files in a single XML file
In all cases, it is possible to filter the results using various criteria such as file name, extension and type of sequence (nucleotide or protein). The tool works even if the files are in distinct sub-folders of the selected folder from which you want to retrieve the files. The procedure is similar to that of "Join folders", but instead of working with folders, in this case the tool manages files permitting the use of key terms as including or excluding filter options to export all files fitting to this term within a single folder or to create a single database file encompassing the contents of all files fitting the filter option. Figure 10.2 provides a screenshot of the Join files process.
|
Figure 10.2. Join Files; 1) Drag your input folder from which you want to retrieve and reorganize contents from the Directory to the "Input folder" box; 2) Check the option, in this case "Select files and place them in a single folder"; 3) type filter; 4) Drop the output folder; and finally run the script. Alternatively, should you select the option "Join files in a single file" the tool will collect the contents of all files and will group them in a single file. In other words, this second option is an easy way to create DNA and protein databases. |
Split files
The script “Split files” is the third tab available at the top of the "Files and folders" GUI and allows you to split a fasta file into different files using the labels of sequences within the original file as classificatory criterion. The options offered by this tool are the following:
- Split files by sequence names
- Split files into blocks
The selection of the sequences is performed over the fasta header according to a key term in the name of the sequences (one or more) or alternatively by blocks of sequences (1000 sequences, 10000 sequences, etc). As shown in Figure 10.3, to use “Split files” drag your input into the corresponding text box (as in the other scripts). Then select one of the proposed split options. For instance, if you want to split your file according to distinct search terms (for instance enzyme acronyms) type each term in the box “Split by sequence names” and then add it to the list at left (you can add as many terms as desired). In doing so, the script will parse the file and will divide it in as many files as search terms used, each one containing the sequences whose fasta labels matched with the term giving the new file its name. The script will also deliver an additional file (no match) containing the remaining sequences of the processed database.
|
Figure 10.3. Split files screnshot. |
Find sequences
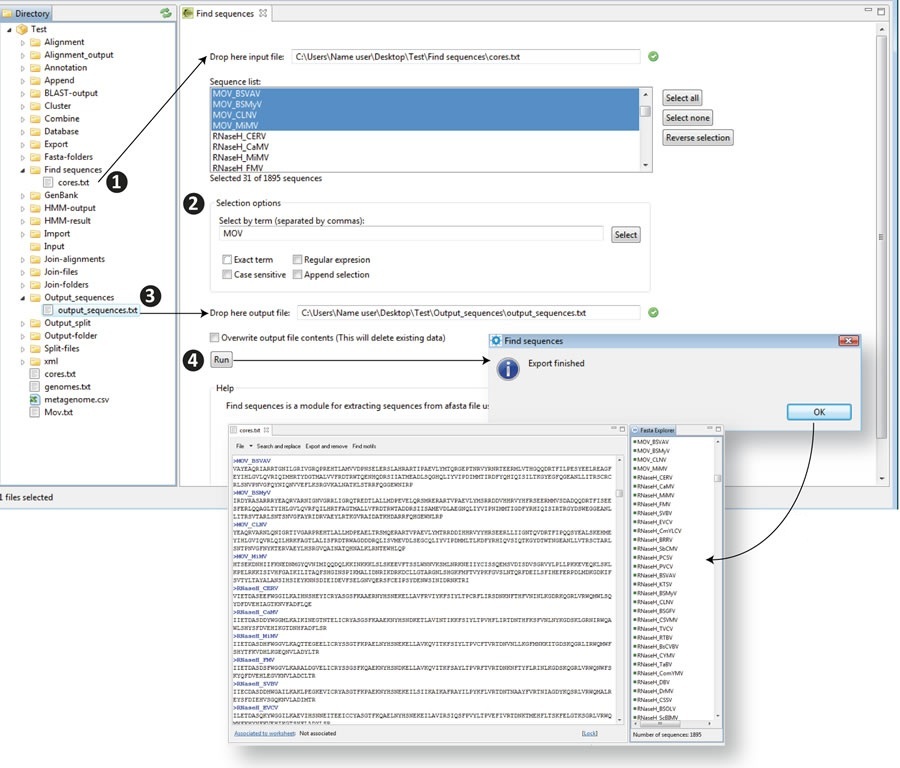
This is a data mining tool for searching and extracting sequences from FASTA file databases using the labels or names of the sequences in the FASTA header as a search criterion. With this utility you can search a fasta file using one or more term options to export sequences and create new databases as described in Figure 10.4.
In this task:
- 1) Drag the input file from the Directory to the "Input file" text box. The FASTA headers of all sequences within the file will be listed in the sequence list dialog
- 2) Type the label or name of those sequences you want to extract and the filter options "Exact term", "Case sensitive", "Regular expresion", "Append selection" or none (which is the default filter) in the box "Select by term” below using a search term or two or more terms by typing them separated by commas. If your file contains any item matching your search terms they will be automatically highlighted in blue within the "Sequence list" dialog (note that to the left of the sequence list you have additional tabs to select all sequences, to deselect them or for reverse the process of selection).
- 3) Drop an output file to the corresponding dialog and choose if you want to overwrite the output file. If you do not make this selection the sequences will be added to the output file without removing previous contents.
- 4) Click "Run" to retrieve and export your selection into a new database within the directory.
|
Figure 10.4. Find sequences script |
Alignments
Frequent jobs in multiple alignment methodology is changing the format of an alignment or trying to identify the set of motifs common to all aligned sequences. GPRO implements two scripts for doing these tasks using DNA/RNA and protein multiple alignment inputs.
Join alignments
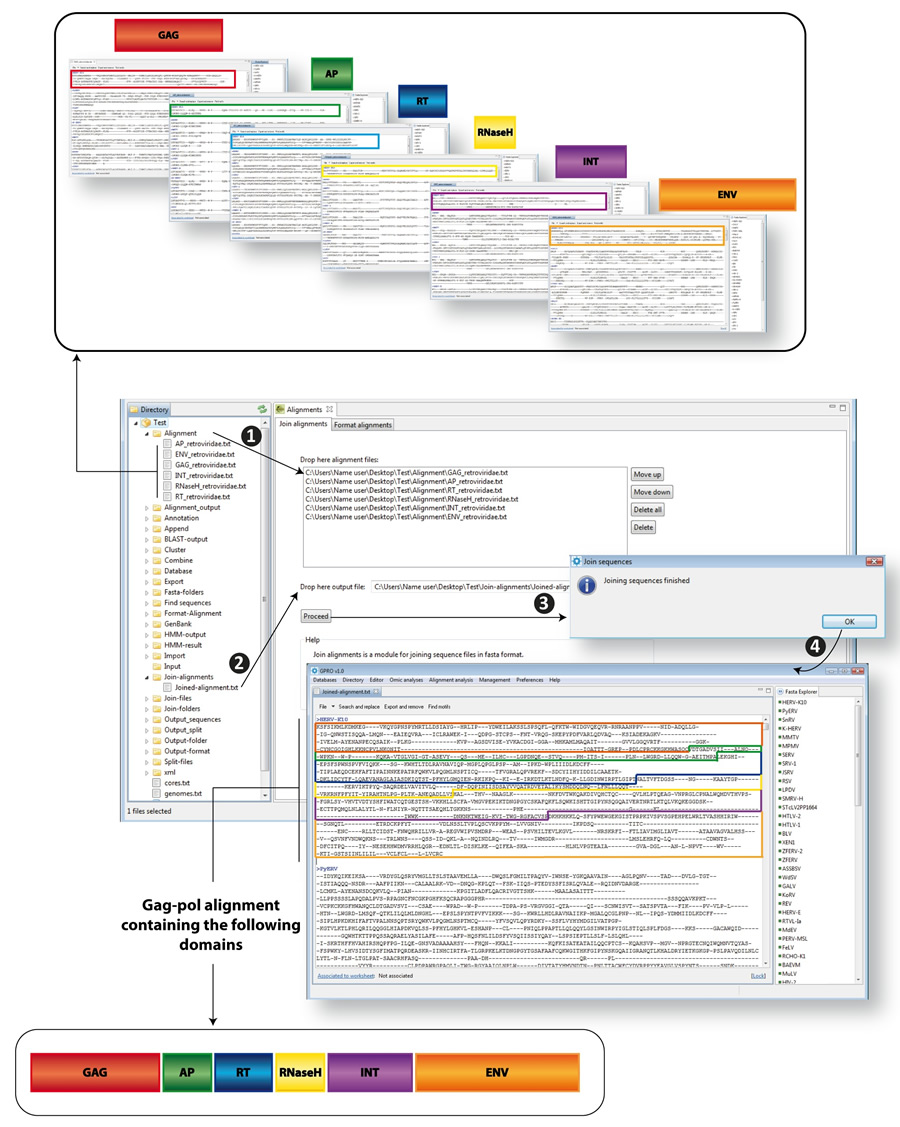
GPRO includes a Join Alignments script allowing you to automatically join different files containing multiple distinct alignments (one per each domain) into a single alignment within a single file and arrange them in a user-defined order. The tool has two requisites. The number and name of the sequences must be identical in each file to join them, and the alignments must be provided in FASTA format. Figure 10.5 illustrates the join alignment process. As usual the process is the followings:
- 1) Drop the alignment files to be joined to the corresponding text box dialog. This dialog lists the distinct files defining the order in which the alignments will be joined. This order can be modified (or removed) using the commands at the right "move up" or "move down"
- 2) Drop an output file into the corresponding dialog
- 3) run the script
- 4) If the number and name of the sequences are identical, the tool will successfully join the sequences and report a single FASTA alignment with all gag-pol domains joined in the specified order for each common name
| Figure 3.24. Join alignments. As an example, we used different files, each containing a multiple alignment based on the GAG (red), Protease (AP, green), Reverse transcriptase (RT, blue), Ribonuclease H (RNaseH, yellow), Integrase (INT, violet) and Envelope (ENV, orange) proteins encoded by distinct Retroviridae retroviruses. We will join these six alignments in a single gag-pol alignment, organized as described in the figure.. |
Format alignments
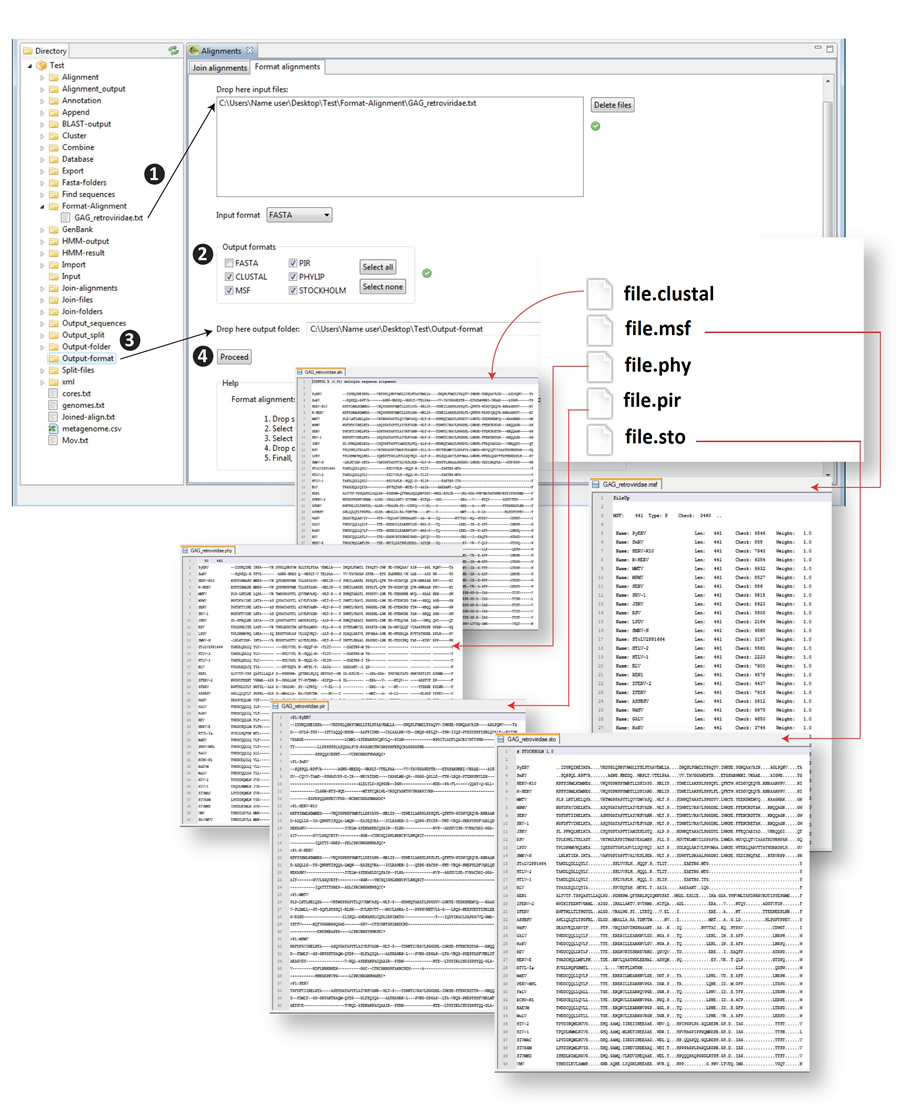
This is an alignment format converter a tool that allows users to upload a protein or nucleotide multiple alignment file in one format and convert it into other formats in one step. The utility accepts and converts the following formats: FASTA, Clustal, Pir, MSF, Phylip and Stockholm. Any of the formats can be used for input and output, and can be selected in one, several or all formats, simultaneously. Figure 10.6 shows a scheme of the procedure.
- 1) Drag the file to be processed from the Directory to the input text box
- 2) Select the format of the input alignment and the format you wish to change it to (aln, msf, phy, pir, sto)
- 3) Drop an output folder into the corresponding dialog
- 4) run the script
|
Figure 10.6. Alignment format converter. |