Description
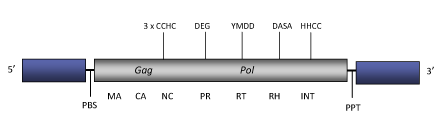
Tamy is a LTR retrotransposon characterized in the saturniid silkworm Antheraea mylitta (Ghosh et al. 2005). Tamy belongs to Pao clade (Copeland et al. 2005) within Branch 2 of the Bel/Pao family (Llorens et al. 2009). Tamy genome is about of 8.4 Kb in size (8387 bp long). Its LTRs of 1305 nt bind the internal coding region that displays a Primer Binding Site (PBS) for a tRNATyr, a single ORF (of 1919 amino acids) for gag and pol domains and a Polypurine Tract (PPT) adjacent to the 3´LTR. The gag nucleocapsid protein contains three zinc finger motifs of Cys-His type typical of Pao-like elements, while pol presents the typical PR, RT, RH and INT domains (Ghosh et al. 2005).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|