Element:TaBV
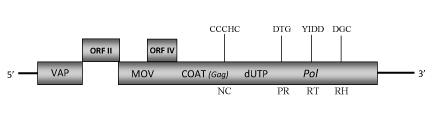
DescriptionTaro (Colocasia esculenta) for its edible corm is one of the most important crop grown throughout many Pacific Island countries. A plant pararetrovirus, Taro bacilliform virus (TaBV), has been isolated from “Alomae” infected taros of Papua New Guinea (PNG) and entirely characterized (Yang et al. 2003a). It is a member of genus Badnavirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. (2009) is located within Class 2 of the Caulimoviridae family. TaBV has bacilliform virions of approximately 125–130 nm x 25–40 nm containing a dsDNA genome that encodes for three Open reading frames (ORFI, II, III) which size and organization are similar to those of other badnaviruses (Yang et al. 2003a). The putative ORF I protein of 146 amino acids shows sequence similarity to the caulimovirus virion associated proteins (VAPs); ORF II potentially encodes a protein of 15.8 kDa which function is still known, while the amino acid sequence of ORF III displays motifs that are highly conserved amongst badnavirus proteins [the movement protein, the COAT zinc finger-like RNA-binding motif (C-X-C-X2-C-X4-H-X4-C) and the second large cysteine-rich region, the aspartic protease, the reverse transcriptase and the RNase H (Yang et al. 2003a)]. The large region rich in zinc finger (CCHC) array duplications (only observed in Badna- and Tungroviruses) presents in the COAT gag-like region of this element resembles to those identified in the nucleocapsid domains of LTR retroelements (Llorens et al. 2009). Interestingly, the TaBV ORF III-polyprotein contains an additional dUTPase domain downstream to the COAT domain similar to that of several Retroviridae retroviruses (Elder et al. 1992) and several Ty3/Gypsy LTR retrotransposons (Novikova and Blinov 2008). A similar dUTPase domain has also been found in the genome of other badnavirus, Dioscorea bacilliform virus (DBV). An additional small ORF (ORF IV) coding for a putative protein of 111 aa is also present, its location within ORF III is similar to that of ORF IV and ORF X of the atypical badnaviruses Citrus yellow mosaic virus (CMBV) and Cacao swollen shoot virus (CSSV), respectively (Yang et al. 2003a). In recent years taros production has suffered a significant decrease due to the severity of the diseases that affect this species. Several viruses have been reported from taro but the most economically significant lethal virus disease is known as "Alomae" (Yang et al. 2003b). It seems to be caused by a double virus infection, indeed into the infected plants have been both isolated Taro bacilliform virus (TaBV) and Colocasia bobone disease virus (CBDV), a putative member of the genus Rhabdovirus (James et al. 1973; Jackson 1978). Has been observed that infection of taro with CBDV alone, known as "bobone disease" (only reported from PNG and the Solomon Islands), determines stunting, leaf distortion and presence of galls on the petioles, whereas the infection with TaBV alone produces mild symptoms including stunting, vein clearing, mosaic and down-curling of the leaf blades (Jackson 1978). Sequence variability in two regions (C-terminal CP and RT/RH) of the genome of South Pacific TaBV isolates has been demonstrated by Yang et al. (2003b). In their study moreover has been detected the presence of a TaBV-like sequence in symptomatic, symptomless and TaBV-free tested plants. This sequence revealed nucleotide similarity to that of TaBV, but it is not yet known whether it is integrated into the taro genome or if it represents the RT/RH-coding region of an additional badnavirus infecting taro (Yang et al. 2003b). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||