Element:SVBV
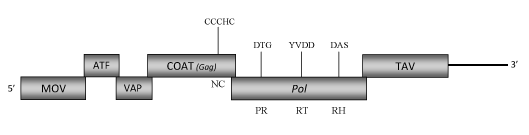
DescriptionStrawberry vein banding virus (SVBV) is a plant pararetrovirus probably distributed world-wide on cultivated strawberries in which it induces the "Strawberry vein banding disease" (Frazier 1955; Morris et al. 1980; Petrzik et al 1998). The virus is transmitted by grafting as well as by aphids in a semi-persistent manner (Frazier 1955). SVBV is a member of the genus Caulimovirus (Caulimoviridae family) (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). SVBV has spherical virions of approximately 40-50 nm in diameter (Kitajima et al. 1973; Morris et al. 1980) composed of a genomic circular double-stranded DNA (7876 bp long) that contains one single-stranded discontinuity typical of the caulimoviruses on each DNA strand. SVBV genome encodes six Open reading frames (ORFs) designated as I, II, III, IV, V and VI (Petrzik et al. 1998). ORF I encodes a putative protein of 350 amino acids (aa) associated to the cell-to-cell movement (MOV). ORF II and III potentially encode two proteins of 161 and 119 aa respectively, whose sequences are similar to those of the "aphid transmission factor" (ATF) and the "virion associated protein" (VAP) usually encoded respectively by the ORFs II and ORFs III of other Caulimoviruses. ORF IV is one of the more conserved genes among Caulimoviruses and contains the "RNA-binding" motif characteristic of the Caulimoviridae COAT (gag)-like domain (C-X-C-X2-C-X4-H-X4-C ) (Hull 1996; Bouhida et al. 1993; Llorens et al 2009). The predicted product of ORF V is a polyprotein of 749 aa displaying the aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains typical of Caulimovirus pol genes. ORF VI, another gene conserved in almost all caulimoviruses, encodes a putative "Translational transactivator protein" (TAV) of 514 aa long (Petrzik et al. 1998). A strong, constitutive SVBV promoter is located in the 3′ terminus of ORF VI and extends downstream into the large intergenic region (Wang et al. 2000; Pattanaik et al. 2004). Its expression activity has been demonstrated to be greater than that of the CaMV 35S promoter, suggesting it might be have a great application in expression of foreign genes in plants (Pattanaik et al. 2004). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||