Element:REV
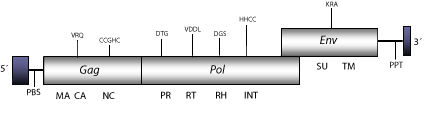
DescriptionAvian Reticuloendotheliosis Virus (REV) is a C-type retrovirus originally described in turkeys and chickens and has also been described as an integrated in the Fowlpox virus genome. REV induces reticuloendotheliosis, a neoplastic disease of the lymphoreticular system in birds (Theilen, Zeigel and Twiehaus 1966; Franklin, Maldonado and Bose 1974; Hoelzer et al. 1980), and carries an oncogene of 1.4 Kb integrated within the env gene (Stephens et al. 1983). During the infection the virus generates two species of unintegrated linear DNA genomes of 8.3 and 5.5 Kb, which represent the virus collaborator (REV-A) and the defective oncogenic component (REV-T), respectively (Chen et al. 1981; Wilhelmsen, Eggleton and Temin 1984). The sequence taken as a representative to describe the genomic structure of REV, is 8 Kb in size including LTRs of 511 and 250 nt, respectively. The internal region displays a Primer Binding Site (PBS) complementary to a tRNAPro, Open Reading Frames (ORFs) for gag-pol, and env genes characteristic of retroviruses, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Hertig et al. 1997; Moore et al. 2000; Kim and Tripathy 2001; Singh, Schitzlein and Tripathy 2003; Garcia et al. 2003; Tadese and Reed 2003). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||