Description
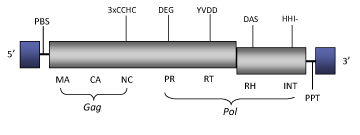
Ninja is a LTR retrotransposon characterized in the insect Drosophila simulans (Ogura et al. 1996). Ninja belongs to Pao clade (Copeland et al. 2005) within Branch 2 of the Bel/Pao family (Llorens et al. 2009). The genome of Ninja is about of 6.6 Kb in size (6644 bp long), including LTRs of 316 nt. The internal region of this LTR retrotransposon displays a Primer Binding Site (PBS), complementary to a tRNASer found in D. melanogaster (Inouye et al. 1986), two ORFs respectively of 1360 and 588 amino acids containing gag- and pol-like regions and a Polypurine Tract (PPT) adjacent to the 3´LTR (Ogura et al. 1996). The largest ORF1 contains both potential gag and pol domains: three putative Cys-motifs characteristic of the gag associated nucleocapsid domain of Bel/Pao LTR retrotransposons and the pol protease and reverse transcriptase domains. The smallest ORF2 contains the putative RNase H and partial integrase pol domains (Ogura et al. 1996; Llorens et al. 2009).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|