Element:KTSV
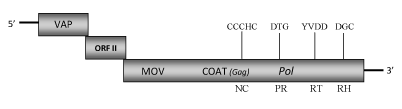
DescriptionKalanchoë top-spotting virus (KTSV) is a plant pararetrovirus isolated from the flowering pot plants Kalanchoë blossfeldiana (Yang et al. 2005). It is a member of the genus Badnavirus of Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005) and according to Llorens et al. 2009 it is located within Class 2 of the Caulimoviridae family. KTSV bacilliform-shaped particles, of approximately 20-25nm x 40-100nm in size (Hearon and Locke 1984), contain a circular double-stranded DNA genome 7591 bp long encoding for three Open reading frames (ORFs I, II, III) which potentially encoded proteins have molecular masses similar to those of the corresponding proteins of other badnaviruses (Yang et al. 2005). The putative ORF I protein shows sequence similarity to the Caulimoviridae virion associated proteins (VAPs). ORF II potentially encodes a protein of 13.8 kDa which function is still unknown. As reported for other badnaviruses, the ORF III product is a polyprotein with the typical cell-to-cell movement protein (MOV), viral gag-like coat (COAT), aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains. The COAT region includes a large region (only observed in Badna- and Tungroviruses) that is rich in zinc finger (CCHC) arrays similar to those found in the LTR retroelement gag-nucleocapsids (Hull 1996; Bouhida et al. 1993; Llorens et al. 2009). KTSV is the causal agent of the "top-spotting disease" of kalanchoë (Hearon and Locke 1984; Lockhart and Ferji 1988). The disease occurs in many commercially traded cultivars of Europe and North America and is characterized by the appearance of numerous chlorotic spots and ring spots on emerging leaves during periods of vigorous vegetative growth. This pararetrovirus can be transmitted by the citrus mealybug vector Planococcus citri, by mechanical inoculation (less) as well as by graft, seed, and pollen (Hearon and Locke 1984). However the natural spread of the top-spotting disease in commercially grown kalanchoë remains primarily by vegetative propagation of infected plants (Yang et al. 2005). KTSV occurs frequently in mixed infections with the Carlavirus sp., Kalanchoë latent virus (KLV, Hearon 1982) and the Potyvirus sp., Kalanchoë mosaic virus (KMV, Husted et al. 1994), (Yang et al. 2005). As found in other badna- and caulimoviruses genome (Harper et al. 2002), the presence of KTSV Endogenous pararetrovirus (EPRV)-like sequences has been also described indicating that they may represent integrated pararetroviral elements in the kalanchoë genome (Yang et al. 2005). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||