Element:EIAV
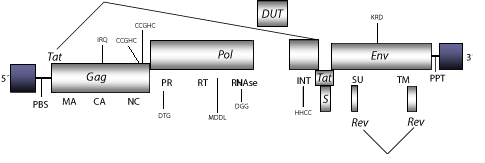
DescriptionEquine Infectious Anemia Virus (EIAV) is a complex lentivirus that induces in horses a disease known as swamp fever or Equine Infectious Anemia (EIA). This disease was originally described in 1900 and is transmitted mechanically via the mouthparts of biting flies of the genus Stomoxys (horse flies and deer flies). Transmission is frequent in swampy regions during the summer. The infection results in acute symptoms in some animals, and chronic fevers, anemia, edema, and cachexia in others. Horses that survive the first clinical episodes are capable of controlling the viral replication and remain asymptomatic. However, all infected horses including those asymptomatic become carriers and are infectious for life (Center for Food Security and Public Health, College of Veterinary Medicine, Iowa State University (2003) and references therein). EIAV can be detected in infected leukocyte cultures at different infection stages, and can be distinguished from other lentiviruses by its triggering a dynamic and variable rapid disease development (Evans, Carpenter and Sevoian 1984). The genomic structure of EIAV is 8.2 Kb including LTRs of 331 nt. The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNALys3, Open Reading Frames (ORFs) for gag, pol and env genes characteristic of retroviruses, tat and rev(both expanded in two exons), which are two accessory genes common in other lentiviruses, an orf S (orf 2) from which little is know, and adjacent to the 3´LTR the Polypurine Tract (PPT) common to other LTR retroelements (McGuire et al. 2000). As other non-primate lentiviruses, EIAV also displays between the RNase H and INT domains an ORF coding for a dUTPase similar to that observed in betaretroviruses, N-terminal to the PR domain ( Elder et al. 1992 ; Payne and Elder 2001 and references therein). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||