Element:CoDi2.4
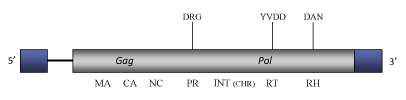
DescriptionCoDi2.4 is a Ty1/Copia LTR retrotransposon described in the genome of the marine pinnate diatom Phaeodactylum tricornutum (Maumus et al. 2009; Bowler et al. 2008). This element belongs to CoDi-I clade (Maumus et al. 2009), within Branch 1 of the Ty1/Copia family (Llorens et al. 2009). Interestingly, CoDi-I represents the first described set of Ty1/Copia elements carrying a putative chromodomain at the C-terminus of the pol-INT domain (Llorens et al. 2009) similarly to Ty3/Gypsy chromoviruses (Marin and Lloréns 2000), which use this feature for chromatin integration (Gao et al. 2008). The genome of CoDi2.4, of 7396 bp in size, includes LTRs of 614 nts bounding a single long Open Reading Frame (ORF) of 1898 amino acids that contains the gag and pol typical structure of the Ty1/Copia LTR retrotransposons (gag, protease, integrase, reverse transcriptase and ribonuclease H, for more details, see Maumus et al. 2009). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||