Element:CmYLCV
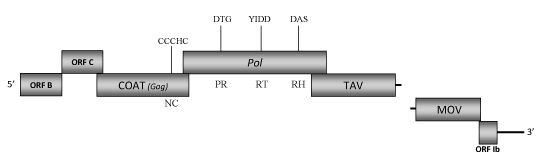
DescriptionCestrum yellow leaf curling virus (CmYLCV), a pararetrovirus closely related to the Soybean chlorotic mottle virus (SbCMV) - the type species of the genus Soymovirus, Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). This virus was first isolated from the green cestrum (Cestrum parqui) a noxious plant (due to its toxicity to livestock, poultry and humans) belonging to the Solanaceae family (Ragozzino 1974). CmYLCV induces pathological alteration on Cestrum parqui plants. The symptoms of infected plants are characterized from yellow mosaic and curling of leaves to shoot stunting (Ragozzino 1974; Stavolone et al. 2003a). Although no insect vector transmission has been yet well demonstrated, it has been observed that healthy C. parqui plants transplanted in the vicinity of CmYLCV-naturally infected plants acquired the virus and displayed the typical symptoms, suggesting a possible insect-mediated transmission (Stavolone et al. 2003a). Morphologically, CmYLCVs are isometric virus particles of about 50 nm in diameter with a circular double-stranded DNA genome 8253 bp long (Stavolone et al. 2003a). The genome contains seven putative Open reading frames (ORFs) designated as I, Ib, II, III, IV, V, VI (Hasegawa et al. 1989; Mushegian et al, 1995; Glasheen et al. 2002; Stavolone et al. 2003a). ORF I encodes for a cell-to-cell movement (MOV) protein of 312 amino acids (aa) in size (Stavolone et al. 2003a). ORF Ib (called as Ia in the GenBank accession) is a small protein located upstream to an intergenic region that includes the tRNAMet binding site. ORF II (namely here as ORF B) encodes a protein of 202 aa also preserved in other soymoviruses whose function is still unclear. This ORF contains however a putative double α-helix domain similar to that found in the ORF II of CaMV, which is involved in both aphid transmission factor (ATF) trimerization (Hebrard et al. 2001) and interaction with the product of ORF III (Leh et al. 1999). ORF III (namely here as ORF C) encodes for a protein of 178 aa containg a characteristic coiled–coil domain responsible for tetramerization -a conserved feature of Caulimovirus "virion associated proteins" (VAPs) (Stavolone et al. 2001). However, our sequence analyses indicate that this protein does not show significant similarities with the VAPs of the other Caulimoviridae species, but it is more related with ORFs C of the other soymoviruses (BRRV and SbCMV) and with the ORF III of the Caulimovirus RuFDV, whose functions are still unclear. ORF IV encodes the viral coat protein precursor as suggested by both similarity analyses and the presence of a "RNA-binding" motif (C-X-C-X2-C-X4-H-X4-C ) (Llorens et al. 2009; Hull 1996; Bouhida et al. 1993). ORF V encodes for a pol polyprotein of 643 aa displaying the typical aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains. ORF VI encodes a putative translational transactivator protein (TAV) of 531 aa that contains the conserved sequence also found in the TAV associated genes of other caulimoviruses (Stavolone et al. 2003a; Glasheen et al. 2002; Hasegawa et al. 1989). CmYLCV differs from other Soymovirus members in the absence of the non-essential ORF VII. The intergenic region (Gap) between TAV and MOV is thus longer than the same region of the remaining known soymoviruses (Mushegian et al, 1995; Takemoto and Hibi 2001; Glasheen et al. 2002]. Within this intergenic region a novel functional constitutive and highly expressed promoter has been isolated (Stavolone et al. 2003b). This promoter is highly active in callus, meristems and vegetative and reproductive tissues of Arabidopsis thaliana, Nicotiana tabacum, Lycopersicon esculentum, Zea mays and Oryza sativa (Stavolone et al. 2003b). The expression levels of this promoter is comparable to, or higher than, the three frequently used promoters in agricultural biotechnology: the Cauliflower mosaic virus (CaMV) 35S (Fang et al. 1989; Benfey et al. 1990), the synthetic "super-promoter" (Ni et al. 1995) or the maize Ubi1 (Christensen et al. 1992)]. Thus, the CmYLCV promoter appears to be a useful tool for biotechnology applications and agricultural research (Stavolone et al. 2003b). The sequence of this promoter is available at Syngenta Biotechnology, Inc. (www.syngentabiotechnology.com). Our sequence analyses additionally reveal the presence of an eighth small ORF, located within the Gag-like gene, similar in size and position to the ORF VIII of SbCMV (Hasegawa et al. 1989). StructureFigure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||