Description
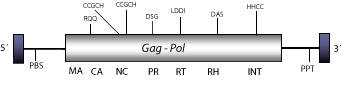
Cigr-1 is a heterogeneous Ty3/Gypsy LTR retrotransposon described in the genome of the urochordate Ciona intestinalis (Simmen et al. 1998). Cigr-1 seems to be transcriptionally active and escapes DNA methylation in C.intestinalis whose genome is a stable mosaic of methylated and nonmethylated domains (Simmen et al. 1999). Although Cigr-1 has been described as an own lineage (Simmen et al. 1999; Simmen and Bird. 2000) its gag polyprotein is strongly similar to those encoded by Mag-like elements. Moreover, gag-pro-pol phylogenetic analyses indicate that Cigr-1 might be a distant Mag recombinant or a sibling lineage of Mag clade (see gag-pro-pol tree in section "phylogenies"). The genomic structure of Cigr-1 is 4.2 Kb in size including LTRs of 245 nt. The internal region of this element displays a single Open Reading Frame (ORF) coding for a gag-pol polyprotein, and a Polypurine Tract (PPT) adjacent to the 3´LTR (Simmen et al. 1998; Simmen and Bird. 2000). Cigr-1 also exhibits, close to the 5´LTR, an unidentified putative Primer Binding Site (PBS) of 18 nt in size characterized by the following "tggccaccataaacatta" DNA motif.
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|