Element:BsCVBV
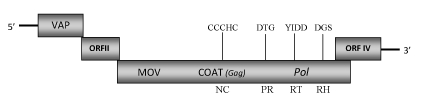
DescriptionBougainvillea is a genus of flowering plants native to South America that worldwide are became a popular ornamental plants in most areas with warm climates. In the species Bougainvillea spectabilis from Taiwan has been recently isolated the pararetrovirus Bougainvillea spectabilis chlorotic vein-banding virus (BsCVBV) (Wang et al. 2007, unpublished) which molecular characteristic resemble to those of Badnavirus genus, Caulimoviridae family (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005). According to Llorens et al. 2009 this still unclassificated badnavirus is located within Class 2 of the Caulimoviridae family. BsCVBV has bacilliform particles that contain a circular dsDNA of about 8.7 Kb in size (8759 bp long) encoding for four Open reading frames (ORFs I, II, III, IV) (Wang et al. 2007, unpublished). ORF I potentially encodes a 15.5 kDa protein showing sequence similarity to the Caulimoviridae virion associated proteins (VAPs) encoded by the corresponding ORFs I. The function of ORF II product (16.4 kDa) is still unknown even if a DNA-binding property has been hypothesized for the ORF II products of some badnaviruses (Borah et al. 2009; Jacquot et al. 1996). The 257.2 kDa polyprotein encoded by ORF III contains the typical Badnavirus cell-to-cell movement protein (MOV), viral coat protein (COAT), aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains. It includes a "RNA-binding" motif sequence "C-X-C-X2-C-X4-H-X4-C" and a second large cysteine-rich region "C-X2-C-X11-C-X2-C-X4-C-X2-C" (only observed in Badna- and Tungroviruses) located at the C-terminus of the COAT (gag)-like region and similar to those identified in the nucleocapsid domains of LTR retroelements (Llorens et al. 2009). ORF IV overlaps the end of ORF III as observed for Citrus yellow mosaic virus (CYMV)-ORF VI and Cacao swollen shoot virus (CSSV)-ORFY (Huang and Hurtung 2001; Hagen et al. 1993). Structure
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||