Element:BRRV
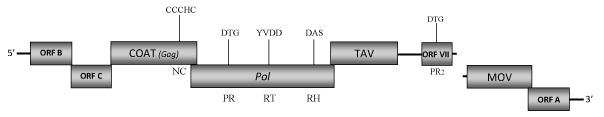
DescriptionBlueberry red ringspot virus (BRRV) is a pararetrovirus member of the genus Soymovirus within the Caulimoviridae (International Committee on the Taxonomy of Viruses -ICTV- Fauquet et al. 2005).This virus is associated with a disease in blueberry plants named red ringspot disease due to the characteristic symptoms that it provokes on leaves and fruits (Hutchinson and Varney 1954; Kim et al. 1981; Ramsdell et al. 1987). Red ringspot disease was first identified in the United States (Hutchinson and Varney 1954), the world’s leading blueberry producer, where has caused multimillionaire losses. The disease was later reported in Europe (Paulechova 1972) and Japan (Isogai et al. 2009). BRRV-affected highbush blueberries (Vaccinium corymbosum) show the typical symptomatology characterized by red ringspots on stems and reddish-brown circular spots of 2–6 mm in diameter on leaves. Furthermore, circular blotches or pale spots can also develop on infected fruit that are often deformed and ripen late (Kim et al.1981; Glasheen et al. 2002; Isogai et al. 2009). BRRV is not mechanically transmitted to herbaceous plants, while is grafting transmissible (Hutchinson and Varney 1954) as well as unknown vectors are involved in its transmission (Isogai et al. 2009). Morphologically, BRRVs are spherical virions of 42-46 nm in diameter that encapsidate a 8303 bp dsDNA genome (Kim et al. 1981). The genome contains eight Open Reading Frames (ORFs) designated as I, A, B, C, IV, V, VI, and VII (Glasheen et al. 2002). ORF I (MOV) encodes a putative protein of 142 amino acids (aa) whose function is associated to cell-to-cell movement. ORFs A and B encode for two unknown proteins of respectively 122 and 186 aa. Functional studies based on these two were performed using other soymoviruses such as Peanut chlorotic streak caulimovirus (PSCV) and Soybean chlorotic mottle virus (SbCMV), which preserve these two ORFs. In PSCV, ORFs A was found to be essential for some aspects of virus life cycle while ORF B was dispensable for infection (Mushegian et al. 1995). In SbCMV, ORF II (named here as ORF B) was essential for the virus infectivity suggesting that it do not represent a transmission helper component (Takemoto and Hibi 2001). However an insect vector is still unknown to date, it is thus speculative to hypothesize ORF B plays a role in insect transmission. Regarding ORF C, comparative analyses performed by us show that this protein reveals sequence similarities with the related ORFs C of the other soymoviruses (SbCMV and CmYLCV) and with the ORF III of the Caulimovirus RuFDV. The function of these ORFs has not been yet well defined and their relationship with the "virion associated protein" (VAP) of Caulimoviridae species remains to be demonstrated. The coat protein gene (ORF IV) is one of the more conserved genes among Caulimoviruses, it contains the characteristic plant pararetroviral "RNA-binding" motif that is larger than the related retroelement consensus RNA-binding sequence (C-X2-C-X4-H-X4-C) due to an additional cysteine (Cys) at the -2 position (C-X-C-X2-C-X4-H-X4-C) (Hull 1996; Bouhida et al. 1993). The predicted product of ORF V is a pol polyprotein of 681 aa displaying the aspartic protease (PR), reverse transcriptase (RT) and RNase H (RH) domains. ORF VI encodes a putative translational transactivator protein (TAV) of 428 aa, a major constituent of the inclusion bodies in which a well-conserved "GLADTIY" sequence also identified in other caulimoviruses, has been found (Hasegawa et al. 1989; Glasheen et al. 2002). ORF VII encodes a protein of 142 aa which function remains unknown, even if in PCSV the related ORF VII has been hypothesized to play a role in host pathogen interactions and in the increased symptom severity (Mushegian et al, 1995). In this regard, our analyses reveal that ORF VII may be an aspartic protease (PR2) additional to that conventionally observed in Pol. A similar feature has been also observed in other soymoviruses, the tungrovirus Rice tungro bacilliform virus (RTBV) and the cavemovirus Cassava vein mosaic virus (CsVMV). The presence of a "Gap" between ORF VI (TAV) and ORF VII seems to be a characteristic of Soymovirus as it has also been observed in the genomes of other members of this genus. StructureFigure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
Related literature |
|
|||||||||||||||||||||||||||||||||||||||||