Description
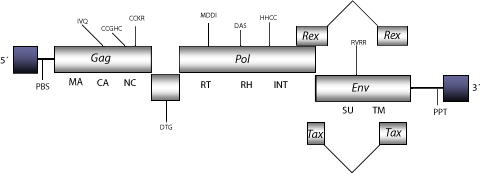
Bovine Leukemia Virus (BLV) is a complex retrovirus associated with Enzootic Bovine Leukosis (EBL), the most common neoplastic cattle disease. Infection by this virus can be clinically silent, but detection and synthesis of BLV virus particles has been observed in infected lymphocyte cultures (Stock and Ferrer 1972; Baliga and Ferrer 1977; Ferrer, Avila, and Stock 1977; Ferrer, Cabradilla and Gupta 1980; Ferrer 1980; Kettmann et al. 1980; Burny et al. 1985; Tajima et al. 1998). Together with other leukemia retroviruses, BLV belongs to the genus Deltaretrovirus (Sagata et al. 1985; Tajima et al. 1998; Fauquet et al. 2005). The genomic structure of BLV is 8.4-8.5 Kb in size including LTRs of 763 nt (pro-5´LTR and pro3´LTR of 321 and 442 nt respectively). The internal region of this retrovirus displays a Primer Binding Site (PBS) complementary to a tRNAPro; three Open Reading Frames (ORFs) for gag, pol, and env genes characteristic of retroviruses, and accessory genes rex and tax, both essential for viral replication and cellular transformation in infected cells (see HTLV-1 or accessory genes), and finally, a Polypurine Tract (PPT) adjacent to 3´LTR (Sagata et al. 1985).
Structure
Figure not to scale. If present, long terminal repeats (LTRs) have been highlighted in blue. Amino acid motifs noted with lines indicate the conserved residues in each protein domain, abbreviations below mean:
| MA=matrix
|
PR=protease
|
DU or DUT=dUTPase
|
TM=transmembrane
|
TAV or IBMP=transactivator/viroplasmin or inclusion body matrix protein
|
| CA=capsid
|
RT=reverse transcriptase
|
INT=Integrase
|
CHR=chromodomain
|
| NC=nucleocapsid
|
RH=RNaseH
|
SU=surface
|
MOV=movement protein
|
| PPT=polypurine tract
|
PBS=primer binding site
|
ATF=aphid transmission factor
|
VAP=virion associated protein
|
Related literature
|
|