CAARD:DDI
Ancestral Maximum Likelihood (ML) Joint Reconstruction
Comments
Ancestral ML reconstructions were performed using FastML 2.02 (Pupko et al. 2000). The tool generates six outputs:
- Pin; reconstructed ancestral NJ tree in newick format.
- Fin; parental relationship among reconstructed nodes and contemporary sequences.
- Jrof; multiple alignment of contemporary sequences and nodes reconstructed with the Joint method.
- Jpf; joint probability per position and total log likelihood.
- Mrof; multiple alignment of contemporary sequences and nodes reconstructed with the marginal method
- Mpf; marginal probability and total log likelihood
Pin output : Ancestral tree
Fin output: Parental relationship among reconstructed nodes and OTUs
| Name | Father | Distance to father | Sons |
| DDIDan | N1 | 0.0262037 | - |
| DDIan | N3 | 0.117206 | - |
| DDIDro | N4 | 0.181222 | - |
| DDISpom | N6 | 0.233917 | - |
| DDIAni | N7 | 0.14889 | - |
| DDINcra | N8 | 0.0910241 | - |
| DDIMgri | N9 | 0.104529 | - |
| DDIGzea | N9 | 0.090694 | - |
| DDIAra | N11 | 0.320097 | - |
| DDIPfal | N13 | 0.0888766 | - |
| DDIPyoe | N13 | 0.0593656 | - |
| DDIGlam | N14 | 0.533159 | - |
| DDICbri | N14 | 0.712107 | - |
| DDILei | N15 | 0.365656 | - |
| DDICer | N15 | 0.477282 | - |
| DDIMfas | N16 | 0.0160301 | - |
| DDIMmus | N16 | 0.0434883 | - |
| DDIXen | N17 | 0.0298944 | - |
| DDIHum | N18 | 0.00328061 | - |
| DDIRnor | N18 | 0.047049 | - |
| N1 | root! | - | DDIDan N2 N17 |
| N2 | N1 | 0.0142114 | N3 N16 |
| N3 | N2 | 0.0822829 | DDIan N4 |
| N4 | N3 | 0.0408853 | DDIDro N5 |
| N5 | N4 | 0.0766323 | N6 N10 |
| N6 | N5 | 0.0943402 | DDISpom N7 |
| N7 | N6 | 0.0413101 | DDIAni N8 |
| N8 | N7 | 0.0307174 | DDINcra N9 |
| N9 | N8 | 0.0126463 | DDIMgri DDIGzea |
| N10 | N5 | 0.0449989 | N11 N15 |
| N11 | N10 | 0.0480625 | DDIAra N12 |
| N12 | N11 | 0.0546856 | N13 N14 |
| N13 | N12 | 0.215828 | DDIPfal DDIPyoe |
| N14 | N12 | 0.201558 | DDIGlam DDICbri |
| N15 | N10 | 0.0204861 | DDILei DDICer |
| N16 | N2 | 0.0304732 | DDIMfas DDIMmus |
| N17 | N1 | 0.00607407 | DDIXen N18 |
| N18 | N17 | 0.0108746 | DDIHum DDIRnor |
Jpf output: Joint probability per amino acid position and total Log Likelihood
| Position | Joint probability | Position | Joint probability | Position | Joint probability | Position | Joint probability |
| 0 | 1.04031e-006 | 28 | 2.07278e-023 | 56 | 3.68473e-015 | 84 | 9.30677e-009 |
| 1 | 0.000458062 | 29 | 6.36391e-006 | 57 | 8.41987e-014 | 85 | 4.35605e-009 |
| 2 | 6.4317e-006 | 30 | 8.78508e-015 | 58 | 1.92041e-010 | 86 | 2.29823e-012 |
| 3 | 0.00362093 | 31 | 0.000266644 | 59 | 5.87121e-016 | 87 | 2.42343e-022 |
| 4 | 0.000266644 | 32 | 2.61792e-014 | 60 | 3.59782e-013 | 88 | 8.1399e-005 |
| 5 | 3.77022e-005 | 33 | 0.00362093 | 61 | 1.87245e-012 | 89 | 4.75546e-005 |
| 6 | 8.74591e-021 | 34 | 2.60946e-005 | 62 | 1.71923e-005 | 90 | 2.80043e-006 |
| 7 | 1.42078e-007 | 35 | 0.00362093 | 63 | 4.77374e-014 | 91 | 2.77581e-018 |
| 8 | 1.56597e-005 | 36 | 1.00043e-015 | 64 | 1.22011e-006 | 92 | 6.87648e-020 |
| 9 | 1.10588e-009 | 37 | 3.74522e-014 | 65 | 1.72592e-012 | 93 | 1.41052e-011 |
| 10 | 1.91651e-006 | 38 | 1.04105e-022 | 66 | 1.85957e-009 | 94 | 1.23053e-013 |
| 11 | 4.02458e-017 | 39 | 0.000821568 | 67 | 0.0129119 | 95 | 1.06902e-015 |
| 12 | 1.14558e-021 | 40 | 1.39619e-005 | 68 | 4.40964e-019 | 96 | 5.59582e-020 |
| 13 | 5.01139e-014 | 41 | 1.88882e-016 | 69 | 5.44922e-015 | 97 | 1.61738e-018 |
| 14 | 0.000266644 | 42 | 0.00362093 | 70 | 2.34288e-021 | 98 | 4.3444e-013 |
| 15 | 2.54724e-010 | 43 | 1.31613e-008 | 71 | 2.69674e-011 | 99 | 0.000358463 |
| 16 | 1.74031e-009 | 44 | 5.87569e-007 | 72 | 2.54187e-008 | 100 | 0.00818293 |
| 17 | 0.00138895 | 45 | 7.05116e-006 | 73 | 7.12699e-017 | 101 | 2.63019e-013 |
| 18 | 1.21429e-014 | 46 | 2.63437e-016 | 74 | 0.0298287 | 102 | 3.82912e-017 |
| 19 | 1.91153e-010 | 47 | 2.68575e-013 | 75 | 4.41244e-007 | 103 | 1.18318e-021 |
| 20 | 2.23125e-015 | 48 | 1.57197e-017 | 76 | 7.64903e-010 | 104 | 5.56663e-024 |
| 21 | 3.22196e-008 | 49 | 1.47113e-012 | 77 | 0.00362093 | 105 | 2.33609e-013 |
| 22 | 3.97195e-007 | 50 | 2.94586e-008 | 78 | 2.3715e-007 | 106 | 8.87572e-022 |
| 23 | 5.30055e-011 | 51 | 8.07382e-009 | 79 | 4.32347e-007 | 107 | 1.98135e-008 |
| 24 | 0.000458062 | 52 | 0.000282579 | 80 | 4.50659e-011 | 108 | 5.63882e-008 |
| 25 | 8.06579e-020 | 53 | 1.00739e-010 | 81 | 0.00429733 | 109 | 6.04427e-017 |
| 26 | 0.000472789 | 54 | 2.78009e-012 | 82 | 5.37664e-012 | ||
| 27 | 5.69615e-016 | 55 | 0.0148872 | 83 | 4.60951e-009 | ||
| Total log likelihood of joint reconstruction: | -2581.74 | ||||||
Jrof output: Ancestral ML Reconstruction Alignment
There are two methods of ancestral reconstruction - Joint and Marginal. In this section, we provide a multiple alignment including both input peptidases and ancestral ML sequences reconstructed using the Joint method.The alignment is available in several formats clicking below the option "Set 1". To build HMM profiles and MRC sequences we removed non-informative amino acid stretches and gaps from several ancestral ML reconstruction analyses You can also retrieve the processed Jrof output, clicking below the option "Set 2". Note however that should you cannot select option 2 is because the output was not processed. <align id="mag" folder="jrof"></align>
Mpf output: Marginal probability per amino acid position and total Log Likelihood
| Position | Joint probability | Position | Joint probability | Position | Joint probability | Position | Joint probability |
| 0 | 1.26812e-006 | 28 | 1.13383e-022 | 56 | 2.32768e-014 | 84 | 1.57364e-008 |
| 1 | 0.00046189 | 29 | 6.47861e-006 | 57 | 1.29608e-013 | 85 | 8.1792e-009 |
| 2 | 1.08606e-005 | 30 | 3.93978e-014 | 58 | 1.09949e-009 | 86 | 4.92287e-012 |
| 3 | 0.00362329 | 31 | 0.000268874 | 59 | 8.92643e-016 | 87 | 6.45839e-022 |
| 4 | 0.000268874 | 32 | 8.39643e-014 | 60 | 5.65055e-013 | 88 | 8.87457e-005 |
| 5 | 3.9447e-005 | 33 | 0.00362329 | 61 | 3.14878e-012 | 89 | 4.81426e-005 |
| 6 | 7.18311e-020 | 34 | 5.28422e-005 | 62 | 1.90804e-005 | 90 | 3.0513e-006 |
| 7 | 2.44681e-007 | 35 | 0.00362329 | 63 | 8.00699e-014 | 91 | 6.92704e-018 |
| 8 | 1.69084e-005 | 36 | 2.80447e-015 | 64 | 1.42404e-006 | 92 | 1.41283e-019 |
| 9 | 2.57643e-009 | 37 | 1.70641e-013 | 65 | 8.43459e-012 | 93 | 1.72979e-011 |
| 10 | 2.08475e-006 | 38 | 3.53471e-021 | 66 | 2.2871e-009 | 94 | 9.09753e-013 |
| 11 | 1.96616e-016 | 39 | 0.00403993 | 67 | 0.06183 | 95 | 5.20779e-015 |
| 12 | 2.19181e-020 | 40 | 1.54558e-005 | 68 | 2.74916e-018 | 96 | 4.09834e-019 |
| 13 | 1.17508e-013 | 41 | 2.73233e-016 | 69 | 5.53363e-014 | 97 | 3.11343e-018 |
| 14 | 0.000268874 | 42 | 0.00362329 | 70 | 3.43617e-020 | 98 | 4.48636e-012 |
| 15 | 2.75739e-010 | 43 | 2.21116e-008 | 71 | 1.02153e-010 | 99 | 0.000547417 |
| 16 | 3.27989e-009 | 44 | 7.10929e-007 | 72 | 5.58255e-008 | 100 | 0.0248586 |
| 17 | 0.001389 | 45 | 7.14359e-006 | 73 | 8.93829e-016 | 101 | 2.41183e-012 |
| 18 | 1.39477e-014 | 46 | 8.81337e-016 | 74 | 0.091904 | 102 | 1.23533e-016 |
| 19 | 5.67625e-010 | 47 | 1.04119e-012 | 75 | 5.68261e-007 | 103 | 5.0654e-021 |
| 20 | 3.94517e-015 | 48 | 8.13928e-017 | 76 | 3.013e-009 | 104 | 8.89264e-024 |
| 21 | 6.78794e-008 | 49 | 2.90448e-012 | 77 | 0.00362329 | 105 | 5.03653e-013 |
| 22 | 1.30172e-006 | 50 | 3.11211e-008 | 78 | 4.16717e-007 | 106 | 6.5902e-021 |
| 23 | 1.1207e-010 | 51 | 1.57966e-008 | 79 | 4.63536e-007 | 107 | 4.42726e-008 |
| 24 | 0.00046189 | 52 | 0.000999038 | 80 | 1.23649e-010 | 108 | 9.17878e-008 |
| 25 | 1.40591e-019 | 53 | 1.10097e-010 | 81 | 0.00430733 | 109 | 2.92644e-016 |
| 26 | 0.000475125 | 54 | 6.5623e-012 | 82 | 1.01129e-011 | ||
| 27 | 6.50203e-016 | 55 | 0.06183 | 83 | 9.90154e-009 | ||
| Total log likelihood of joint reconstruction: | -2490.02 | ||||||
Mrof output: Ancestral ML Reconstruction Alignment
There are two methods of ancestral reconstruction - Joint and Marginal. In this section, we provide a multiple alignment including both input peptidases and ancestral ML sequences reconstructed using the Joint method.The alignment is available in several formats clicking below the option "Set 1". To build HMM profiles and MRC sequences we removed non-informative amino acid stretches and gaps from several ancestral ML reconstruction analyses You can also retrieve the processed Jrof output, clicking below the option "Set 2". Note however that should you cannot select option 2 is because the output was not processed. <align id="mag" folder="mrof"></align>
Models
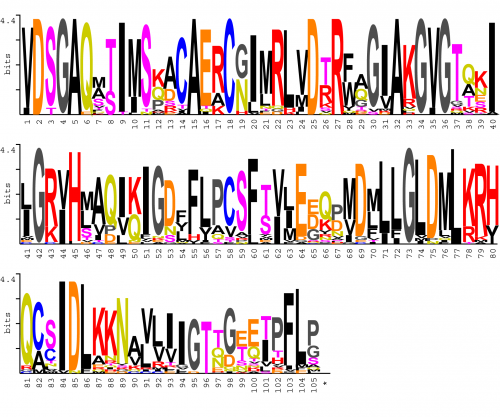
Sequence Logo
Sequence logo constructed from the input of the processed Jrof alignment. In every position, each residue is a letter whose height is proportional to its frequency multiplied by the information content of each position measured in bits. Letters are placed such that the most common is at the top.
- Basic residues are represented in red
- Hydrophobic residues in black
- Amino acids frequent in β-turns (G and P) in dark grey
- Small nucleophiles in violet
- Acidic residues in orange
- Acidic-relative amides in green
The logo was constructed using ChekAlign server with the Shannon's algorithm (Shannon 1997) and options "include gaps" and "Correction factor". Gaps are not represented by any symbol but occupy a blank also proportional to its frequency and, for aesthetic reasons, always at the top. Maximum entropy is log221. The alignment gap is considered to be another state or amino acid species.
HMMs
>AP_ddi profile HMM generated consensus sequence VDSGAQatImSkacAErCGimRLvDtRfqGiAkGVGtqkIlGrIHlaqikiGdFLpcsF tVlEdqpmDlLLGLDmLkrHQccIDLkknllitgeetpFLp
Pairwise alignment between the MRC sequence and the DTG/ILG template HMM-profile
Domain 1 of 1, from 1 to 85: score 59.2, E = 1.5e-18
DTG_ILG template *->vDTGAsvlsviskecklaqklgltrkkafdpSSYvCivtllsysqPs
vD+GA+ +++sk c a+++g + ++ +
AP_ddi 1 VDSGAQA-TIMSKAC--AERCGIM-------------------RLVD 25
sktsttaqdtirgagGqskiyvSklktsgqirknllslvtikitkGnvTe
+ ++ +a+G + k++g+i+ l iki G
AP_ddi 26 T-RF------QGIAKGVGTQ-----KILGRIH-----LAQIKI--G---- 52
venrslpsdgvflvv.tdpedqksrydvILGrldfLrqlnsvhidl<-*
lp++ f v++++p +d++LG+ d+L+ ++++ idl
AP_ddi 53 ---DFLPCS--FTVLeDQP------MDLLLGL-DMLKRHQCC-IDL 85
Cite this site
Llorens, C. Futami, R. Renaud, G. and A. Moya (2009). Bioinformatic Flowchart and Database to Investigate the Diversity of Clan AA Peptidases.Biology Direct, 4:3.